First steps in pandas#
This very short introduction to pandas is mainly based on the excellent pandas documentation.
To use this tutorial, I recommend the following procedure:
On your machine, create a new folder called
pandasDownload this tutorial as .ipynb (on the top right of this webpage, select the download button) and move it to your
pandasfolderOpen Visual Studio Code and select the “Explorer” symbol on the top left in the Activity Bar
Select “Open Folder” and choose your folder
pandas. This folder is now your project directoryIn the Explorer, open the file
pandas-intro-short.ipynb
Import pandas#
To load the pandas package and start working with it, import the package.
The community agreed alias for pandas is
pd, so loading pandas as pd is assumed standard practice for all of the pandas documentation:
import pandas as pd
Data creation#
To manually store data in a table, create a DataFrame:
# create the DataFrame and name it my_df
my_df = pd.DataFrame(
{
'name': [ "Tom", "Lisa", "Peter"],
'height': [1.68, 1.93, 1.72],
'weight': [48.4, 89.8, 84.2],
'gender': ['male', 'female', 'male']
}
)
# show my_df
my_df
| name | height | weight | gender | |
|---|---|---|---|---|
| 0 | Tom | 1.68 | 48.4 | male |
| 1 | Lisa | 1.93 | 89.8 | female |
| 2 | Peter | 1.72 | 84.2 | male |
Import data#
pandas supports many different file formats or data sources out of the box (csv, excel, sql, json, parquet, …)
each of them import data with the prefix
read_*Import data, available as a CSV file in a GitHub repo:
df = pd.read_csv("https://raw.githubusercontent.com/kirenz/datasets/master/height_unclean.csv", delimiter=";", decimal=",")
# show head
df.head()
| Name | ID% | Height | Average Height Parents | Gender | |
|---|---|---|---|---|---|
| 0 | Stefanie | 1 | 162 | 161.5 | female |
| 1 | Peter | 2 | 163 | 163.5 | male |
| 2 | Stefanie | 3 | 163 | 163.2 | female |
| 3 | Manuela | 4 | 164 | 165.1 | female |
| 4 | Simon | 5 | 164 | 163.2 | male |
# same import with different style
ROOT = "https://raw.githubusercontent.com/kirenz/datasets/master/"
DATA = "height_unclean.csv"
df = pd.read_csv(ROOT + DATA, delimiter=";", decimal=",")
# show head
df.head()
| Name | ID% | Height | Average Height Parents | Gender | |
|---|---|---|---|---|---|
| 0 | Stefanie | 1 | 162 | 161.5 | female |
| 1 | Peter | 2 | 163 | 163.5 | male |
| 2 | Stefanie | 3 | 163 | 163.2 | female |
| 3 | Manuela | 4 | 164 | 165.1 | female |
| 4 | Simon | 5 | 164 | 163.2 | male |
Store data#
pandas supports many different file formats (csv, excel, sql, json, parquet, …)
each of them stores data with the prefix
to_*The following code will save data as an Excel file in your current directory (you may need to install OpenPyXL first.
In the example here, the
sheet_nameis named people_height instead of the default Sheet1. By settingindex=Falsethe row index labels are not saved in the spreadsheet:
df.to_excel("height.xlsx", sheet_name="people_height", index=False)
The equivalent read function
read_excel()would reload the data to a DataFrame:
# load excel file
df_new = pd.read_excel("height.xlsx", sheet_name="people_height")
Viewing data#
Overview#
# show df
df
| Name | ID% | Height | Average Height Parents | Gender | |
|---|---|---|---|---|---|
| 0 | Stefanie | 1 | 162 | 161.5 | female |
| 1 | Peter | 2 | 163 | 163.5 | male |
| 2 | Stefanie | 3 | 163 | 163.2 | female |
| 3 | Manuela | 4 | 164 | 165.1 | female |
| 4 | Simon | 5 | 164 | 163.2 | male |
| 5 | Sophia | 6 | 164 | 164.4 | female |
| 6 | Ellen | 7 | 164 | 164.0 | female |
| 7 | Emilia | 8 | 165 | 165.2 | female |
| 8 | Lina | 9 | 165 | 165.2 | female |
| 9 | Marie | 10 | 165 | 165.1 | female |
| 10 | Lena | 11 | 165 | 166.3 | female |
| 11 | Mila | 12 | 165 | 167.4 | female |
| 12 | Fin | 13 | 165 | 165.5 | male |
| 13 | Eric | 14 | 166 | 166.2 | male |
| 14 | Pia | 15 | 166 | 166.1 | female |
| 15 | Marc | 16 | 166 | 166.5 | male |
| 16 | Ralph | 17 | 166 | 166.6 | male |
| 17 | Tom | 18 | 167 | 166.2 | male |
| 18 | Steven | 19 | 167 | 167.3 | male |
| 19 | Emanuel | 20 | 168 | 168.5 | male |
# show first 2 rows
df.head(2)
| Name | ID% | Height | Average Height Parents | Gender | |
|---|---|---|---|---|---|
| 0 | Stefanie | 1 | 162 | 161.5 | female |
| 1 | Peter | 2 | 163 | 163.5 | male |
# show last 2 rows
df.tail(2)
| Name | ID% | Height | Average Height Parents | Gender | |
|---|---|---|---|---|---|
| 18 | Steven | 19 | 167 | 167.3 | male |
| 19 | Emanuel | 20 | 168 | 168.5 | male |
The
info()method prints information about a DataFrame including the index dtype and columns, non-null values and memory usage:
df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 20 entries, 0 to 19
Data columns (total 5 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Name 20 non-null object
1 ID% 20 non-null int64
2 Height 20 non-null int64
3 Average Height Parents 20 non-null float64
4 Gender 20 non-null object
dtypes: float64(1), int64(2), object(2)
memory usage: 928.0+ bytes
Column names#
# Show columns
df.columns
Index(['Name', 'ID%', 'Height', 'Average Height Parents', ' Gender'], dtype='object')
Data type#
Show data types (dtypes).
df.dtypes
Name object
ID% int64
Height int64
Average Height Parents float64
Gender object
dtype: object
The data types in this DataFrame are integers (int64), floats (float64) and strings (object).
Index#
# Only show index
df.index
RangeIndex(start=0, stop=20, step=1)
Change column names#
Usually, we prefer to work with columns that have the following proporties:
no leading or trailing whitespace (
"name"instead of" name "," name"or"name ")all lowercase (
"name"instead of"Name")now white spaces (
"my_name"instead of"my name")
Simple rename#
First, we rename columns by simply using a mapping
We rename
"Name"to"name"and just print the result (we want to display errors and don’t save the changes for now):
df.rename(columns={"Name": "name"}, errors="raise")
| name | ID% | Height | Average Height Parents | Gender | |
|---|---|---|---|---|---|
| 0 | Stefanie | 1 | 162 | 161.5 | female |
| 1 | Peter | 2 | 163 | 163.5 | male |
| 2 | Stefanie | 3 | 163 | 163.2 | female |
| 3 | Manuela | 4 | 164 | 165.1 | female |
| 4 | Simon | 5 | 164 | 163.2 | male |
| 5 | Sophia | 6 | 164 | 164.4 | female |
| 6 | Ellen | 7 | 164 | 164.0 | female |
| 7 | Emilia | 8 | 165 | 165.2 | female |
| 8 | Lina | 9 | 165 | 165.2 | female |
| 9 | Marie | 10 | 165 | 165.1 | female |
| 10 | Lena | 11 | 165 | 166.3 | female |
| 11 | Mila | 12 | 165 | 167.4 | female |
| 12 | Fin | 13 | 165 | 165.5 | male |
| 13 | Eric | 14 | 166 | 166.2 | male |
| 14 | Pia | 15 | 166 | 166.1 | female |
| 15 | Marc | 16 | 166 | 166.5 | male |
| 16 | Ralph | 17 | 166 | 166.6 | male |
| 17 | Tom | 18 | 167 | 166.2 | male |
| 18 | Steven | 19 | 167 | 167.3 | male |
| 19 | Emanuel | 20 | 168 | 168.5 | male |
Let`s rename Gender to gender
Again, we just want to display the result (without saving it).
Remove the # and run the following code:
# df.rename(columns={"Gender": "gender"}, errors="raise")
This raises an error. Can you spot the problem? (take a look at the end of the error statement)
The KeyError statement tells us that
"['Gender'] not found in axis"This is because variable Gender has a white space at the beginning:
[ Gender]We could fix this problem by typing
" Gender"instead of"Gender"However, there are useful functions (regular expressions) to deal with this kind of problems
Trailing and leading spaces (with regex)#
We use regular expressions to deal with whitespaces
To change multiple column names at once, we use the method
.columns.strTo replace the spaces, we use
.replace()withregex=TrueIn the following function, we search for leading (line start and spaces) and trailing (spaces and line end) spaces and replace them with an empty string:
# replace r"this pattern" with empty string r""
df.columns = df.columns.str.replace(r"^ +| +$", r"", regex=True)
Explanation for the regex (see also Stackoverflow):
we start with
r(for raw) which tells Python to treat all following input as raw text (without interpreting it)“
^”: is line start“
+”: (space and plus) is one or more spaces“
|”: is or“
$”: is line end
To learn more about regular expressions (“regex”), visit the following sites:
Replace special characters#
Again, we use regular expressions to deal with special characters (like %, &, $ etc.)
# replace r"this pattern" with empty string r""
df.columns = df.columns.str.replace(r"%", r"", regex=True)
df.columns
Index(['Name', 'ID', 'Height', 'Average Height Parents', 'Gender'], dtype='object')
Lowercase and whitespace#
We can use two simple methods to convert all columns to lowercase and replace white spaces with underscores (“_”):
df.columns = df.columns.str.lower().str.replace(' ', '_')
df.columns
Index(['name', 'id', 'height', 'average_height_parents', 'gender'], dtype='object')
Change data type#
There are several methods to change data types in pandas:
.astype(): Convert to a specific type (like “int32”, “float” or “catgeory”)to_datetime: Convert argument to datetime.to_timedelta: Convert argument to timedelta.to_numeric: Convert argument to a numeric type.numpy.ndarray.astype: Cast a numpy array to a specified type.
Categorical data#
Categoricals are a pandas data type corresponding to categorical variables in statistics.
A categorical variable takes on a limited, and usually fixed, number of possible values (categories). Examples are gender, social class, blood type, country affiliation, observation time or rating via Likert scales.
Converting an existing column to a category dtype:
df["gender"] = df["gender"].astype("category")
df.dtypes
name object
id int64
height int64
average_height_parents float64
gender category
dtype: object
String data#
In our example, id is not a number (we can’t perform calculations with it)
It is just a unique identifier so we should transform it to a simple string (object)
df['id'] = df['id'].astype(str)
df.dtypes
name object
id object
height int64
average_height_parents float64
gender category
dtype: object
Add new columns#
Constant#
# add a constant to all rows
df["number"] = 42
df.head(3)
| name | id | height | average_height_parents | gender | number | |
|---|---|---|---|---|---|---|
| 0 | Stefanie | 1 | 162 | 161.5 | female | 42 |
| 1 | Peter | 2 | 163 | 163.5 | male | 42 |
| 2 | Stefanie | 3 | 163 | 163.2 | female | 42 |
From existing#
Create new column from existing columns
import numpy as np
# calculate height in m (from cm)
df['height_m'] = df.height/100
# add some random numbers
df['weight'] = round(np.random.normal(45, 5, 20) * df['height_m'],2)
# calculate body mass index
df['bmi'] = round(df.weight / (df.height_m * df.height_m),2)
Date#
To add a date, we can use datetime and strftime:
# add date
from datetime import datetime
df["date"] = datetime.today().strftime('%Y-%m-%d')
df.head(3)
| name | id | height | average_height_parents | gender | number | height_m | weight | bmi | date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Stefanie | 1 | 162 | 161.5 | female | 42 | 1.62 | 74.98 | 28.57 | 2023-03-21 |
| 1 | Peter | 2 | 163 | 163.5 | male | 42 | 1.63 | 62.06 | 23.36 | 2023-03-21 |
| 2 | Stefanie | 3 | 163 | 163.2 | female | 42 | 1.63 | 77.19 | 29.05 | 2023-03-21 |
Summary statistics#
Numeric data#
describe() shows a quick statistic summary of your numerical data.
We transpose the data (with
.T) to make it more readable:
df.describe().T
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| height | 20.0 | 165.000 | 1.486784 | 162.00 | 164.000 | 165.00 | 166.0000 | 168.00 |
| average_height_parents | 20.0 | 165.350 | 1.687883 | 161.50 | 164.300 | 165.35 | 166.3500 | 168.50 |
| number | 20.0 | 42.000 | 0.000000 | 42.00 | 42.000 | 42.00 | 42.0000 | 42.00 |
| height_m | 20.0 | 1.650 | 0.014868 | 1.62 | 1.640 | 1.65 | 1.6600 | 1.68 |
| weight | 20.0 | 78.430 | 8.402891 | 62.06 | 74.405 | 79.06 | 83.6925 | 92.04 |
| bmi | 20.0 | 28.806 | 3.049660 | 23.02 | 27.395 | 28.99 | 30.7100 | 33.81 |
Obtain summary statistics for different groups (categorical data)
df.groupby(['gender']).describe().T
| gender | female | male | |
|---|---|---|---|
| height | count | 11.000000 | 9.000000 |
| mean | 164.363636 | 165.777778 | |
| std | 1.120065 | 1.563472 | |
| min | 162.000000 | 163.000000 | |
| 25% | 164.000000 | 165.000000 | |
| 50% | 165.000000 | 166.000000 | |
| 75% | 165.000000 | 167.000000 | |
| max | 166.000000 | 168.000000 | |
| average_height_parents | count | 11.000000 | 9.000000 |
| mean | 164.863636 | 165.944444 | |
| std | 1.593909 | 1.693451 | |
| min | 161.500000 | 163.200000 | |
| 25% | 164.200000 | 165.500000 | |
| 50% | 165.100000 | 166.200000 | |
| 75% | 165.650000 | 166.600000 | |
| max | 167.400000 | 168.500000 | |
| number | count | 11.000000 | 9.000000 |
| mean | 42.000000 | 42.000000 | |
| std | 0.000000 | 0.000000 | |
| min | 42.000000 | 42.000000 | |
| 25% | 42.000000 | 42.000000 | |
| 50% | 42.000000 | 42.000000 | |
| 75% | 42.000000 | 42.000000 | |
| max | 42.000000 | 42.000000 | |
| height_m | count | 11.000000 | 9.000000 |
| mean | 1.643636 | 1.657778 | |
| std | 0.011201 | 0.015635 | |
| min | 1.620000 | 1.630000 | |
| 25% | 1.640000 | 1.650000 | |
| 50% | 1.650000 | 1.660000 | |
| 75% | 1.650000 | 1.670000 | |
| max | 1.660000 | 1.680000 | |
| weight | count | 11.000000 | 9.000000 |
| mean | 81.221818 | 75.017778 | |
| std | 7.257694 | 8.833854 | |
| min | 67.780000 | 62.060000 | |
| 25% | 77.280000 | 67.860000 | |
| 50% | 79.650000 | 77.680000 | |
| 75% | 87.090000 | 82.140000 | |
| max | 92.040000 | 85.480000 | |
| bmi | count | 11.000000 | 9.000000 |
| mean | 30.050909 | 27.284444 | |
| std | 2.490636 | 3.098214 | |
| min | 25.200000 | 23.020000 | |
| 25% | 28.695000 | 24.330000 | |
| 50% | 29.260000 | 27.520000 | |
| 75% | 31.985000 | 29.810000 | |
| max | 33.810000 | 31.400000 |
Categorical data#
we can also use
describe()for categorical data
df.describe(include="category").T
| count | unique | top | freq | |
|---|---|---|---|---|
| gender | 20 | 2 | female | 11 |
Show unique levels and count with
value_counts()
df['gender'].value_counts()
female 11
male 9
Name: gender, dtype: int64
Loop over list#
Example of for loop to obtain statistics for specific numerical columns
# make a list of numerical columns
list_num = ['height', 'weight']
# calculate median for our list
for i in list_num:
print(f'Median of {i} equals {df[i].median()} \n')
Median of height equals 165.0
Median of weight equals 79.06
# calculate summary statistics for our list
for i in list_num:
print(f'Column: {i} \n {df[i].describe().T} \n')
Column: height
count 20.000000
mean 165.000000
std 1.486784
min 162.000000
25% 164.000000
50% 165.000000
75% 166.000000
max 168.000000
Name: height, dtype: float64
Column: weight
count 20.000000
mean 78.430000
std 8.402891
min 62.060000
25% 74.405000
50% 79.060000
75% 83.692500
max 92.040000
Name: weight, dtype: float64
import seaborn as sns
import matplotlib.pyplot as plt
# obtain plots for our list
for i in list_num:
sns.boxplot(x="gender", y=i, data=df)
plt.title("Boxplot for metric " + i)
plt.show()
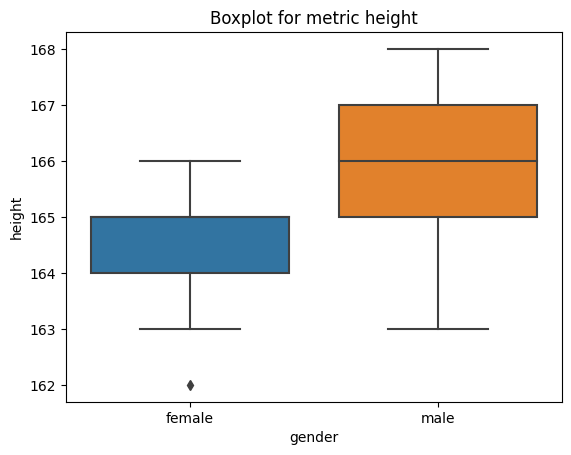
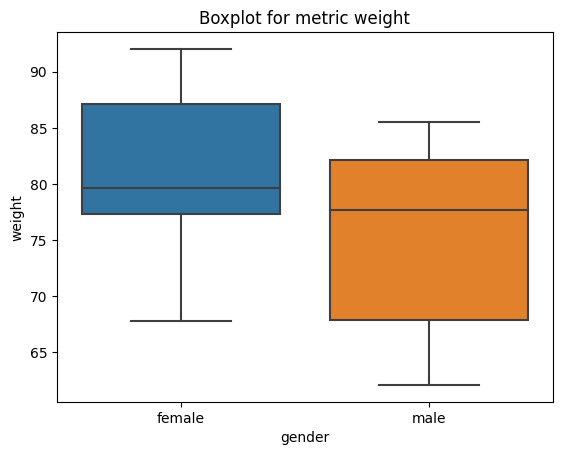
Sorting#
Sorting by values:
df.sort_values(by="height")
| name | id | height | average_height_parents | gender | number | height_m | weight | bmi | date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Stefanie | 1 | 162 | 161.5 | female | 42 | 1.62 | 74.98 | 28.57 | 2023-03-21 |
| 1 | Peter | 2 | 163 | 163.5 | male | 42 | 1.63 | 62.06 | 23.36 | 2023-03-21 |
| 2 | Stefanie | 3 | 163 | 163.2 | female | 42 | 1.63 | 77.19 | 29.05 | 2023-03-21 |
| 3 | Manuela | 4 | 164 | 165.1 | female | 42 | 1.64 | 85.35 | 31.73 | 2023-03-21 |
| 4 | Simon | 5 | 164 | 163.2 | male | 42 | 1.64 | 72.68 | 27.02 | 2023-03-21 |
| 5 | Sophia | 6 | 164 | 164.4 | female | 42 | 1.64 | 67.78 | 25.20 | 2023-03-21 |
| 6 | Ellen | 7 | 164 | 164.0 | female | 42 | 1.64 | 81.99 | 30.48 | 2023-03-21 |
| 12 | Fin | 13 | 165 | 165.5 | male | 42 | 1.65 | 85.48 | 31.40 | 2023-03-21 |
| 11 | Mila | 12 | 165 | 167.4 | female | 42 | 1.65 | 79.65 | 29.26 | 2023-03-21 |
| 10 | Lena | 11 | 165 | 166.3 | female | 42 | 1.65 | 89.79 | 32.98 | 2023-03-21 |
| 9 | Marie | 10 | 165 | 165.1 | female | 42 | 1.65 | 78.47 | 28.82 | 2023-03-21 |
| 8 | Lina | 9 | 165 | 165.2 | female | 42 | 1.65 | 92.04 | 33.81 | 2023-03-21 |
| 7 | Emilia | 8 | 165 | 165.2 | female | 42 | 1.65 | 77.37 | 28.42 | 2023-03-21 |
| 13 | Eric | 14 | 166 | 166.2 | male | 42 | 1.66 | 83.14 | 30.17 | 2023-03-21 |
| 14 | Pia | 15 | 166 | 166.1 | female | 42 | 1.66 | 88.83 | 32.24 | 2023-03-21 |
| 15 | Marc | 16 | 166 | 166.5 | male | 42 | 1.66 | 82.14 | 29.81 | 2023-03-21 |
| 16 | Ralph | 17 | 166 | 166.6 | male | 42 | 1.66 | 63.43 | 23.02 | 2023-03-21 |
| 17 | Tom | 18 | 167 | 166.2 | male | 42 | 1.67 | 67.86 | 24.33 | 2023-03-21 |
| 18 | Steven | 19 | 167 | 167.3 | male | 42 | 1.67 | 80.69 | 28.93 | 2023-03-21 |
| 19 | Emanuel | 20 | 168 | 168.5 | male | 42 | 1.68 | 77.68 | 27.52 | 2023-03-21 |
Selection#
Getting []#
Selecting a single column (equivalent to df.height):
df["height"]
0 162
1 163
2 163
3 164
4 164
5 164
6 164
7 165
8 165
9 165
10 165
11 165
12 165
13 166
14 166
15 166
16 166
17 167
18 167
19 168
Name: height, dtype: int64
Selecting via [], which slices the rows (endpoint is not included).
df[0:1]
| name | id | height | average_height_parents | gender | number | height_m | weight | bmi | date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Stefanie | 1 | 162 | 161.5 | female | 42 | 1.62 | 74.98 | 28.57 | 2023-03-21 |
By label .loc#
The .loc attribute is the primary access method. The following are valid inputs:
For getting a cross section using a label:
df.loc[[0]]
| name | id | height | average_height_parents | gender | number | height_m | weight | bmi | date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Stefanie | 1 | 162 | 161.5 | female | 42 | 1.62 | 74.98 | 28.57 | 2023-03-21 |
Selecting on a multi-axis by label:
df.loc[ : , ["name", "height"]]
| name | height | |
|---|---|---|
| 0 | Stefanie | 162 |
| 1 | Peter | 163 |
| 2 | Stefanie | 163 |
| 3 | Manuela | 164 |
| 4 | Simon | 164 |
| 5 | Sophia | 164 |
| 6 | Ellen | 164 |
| 7 | Emilia | 165 |
| 8 | Lina | 165 |
| 9 | Marie | 165 |
| 10 | Lena | 165 |
| 11 | Mila | 165 |
| 12 | Fin | 165 |
| 13 | Eric | 166 |
| 14 | Pia | 166 |
| 15 | Marc | 166 |
| 16 | Ralph | 166 |
| 17 | Tom | 167 |
| 18 | Steven | 167 |
| 19 | Emanuel | 168 |
Showing label slicing, both endpoints are included:
df.loc[0:1, ["name", "height"]]
| name | height | |
|---|---|---|
| 0 | Stefanie | 162 |
| 1 | Peter | 163 |
Reduction in the dimensions of the returned object:
df.loc[0, ["name", "height"]]
name Stefanie
height 162
Name: 0, dtype: object
For getting a scalar value:
df.loc[[0], "height"]
0 162
Name: height, dtype: int64
By position .iloc#
pandas provides a suite of methods in order to get purely integer based indexing. Here, the .iloc attribute is the primary access method.
df.iloc[0]
name Stefanie
id 1
height 162
average_height_parents 161.5
gender female
number 42
height_m 1.62
weight 74.98
bmi 28.57
date 2023-03-21
Name: 0, dtype: object
By integer slices:
df.iloc[0:2, 0:2]
| name | id | |
|---|---|---|
| 0 | Stefanie | 1 |
| 1 | Peter | 2 |
By lists of integer position locations:
df.iloc[[0, 2], [0, 2]]
| name | height | |
|---|---|---|
| 0 | Stefanie | 162 |
| 2 | Stefanie | 163 |
For slicing rows explicitly:
df.iloc[1:3, :]
| name | id | height | average_height_parents | gender | number | height_m | weight | bmi | date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Peter | 2 | 163 | 163.5 | male | 42 | 1.63 | 62.06 | 23.36 | 2023-03-21 |
| 2 | Stefanie | 3 | 163 | 163.2 | female | 42 | 1.63 | 77.19 | 29.05 | 2023-03-21 |
For slicing columns explicitly:
df.iloc[:, 1:3]
| id | height | |
|---|---|---|
| 0 | 1 | 162 |
| 1 | 2 | 163 |
| 2 | 3 | 163 |
| 3 | 4 | 164 |
| 4 | 5 | 164 |
| 5 | 6 | 164 |
| 6 | 7 | 164 |
| 7 | 8 | 165 |
| 8 | 9 | 165 |
| 9 | 10 | 165 |
| 10 | 11 | 165 |
| 11 | 12 | 165 |
| 12 | 13 | 165 |
| 13 | 14 | 166 |
| 14 | 15 | 166 |
| 15 | 16 | 166 |
| 16 | 17 | 166 |
| 17 | 18 | 167 |
| 18 | 19 | 167 |
| 19 | 20 | 168 |
For getting a value explicitly:
df.iloc[0, 0]
'Stefanie'
Filter (boolean indexing)#
Using a single column’s values to select data.
df[df["height"] > 180]
| name | id | height | average_height_parents | gender | number | height_m | weight | bmi | date |
|---|
Using the isin() method for filtering:
df[df["name"].isin(["Tom", "Lisa"])]
| name | id | height | average_height_parents | gender | number | height_m | weight | bmi | date | |
|---|---|---|---|---|---|---|---|---|---|---|
| 17 | Tom | 18 | 167 | 166.2 | male | 42 | 1.67 | 67.86 | 24.33 | 2023-03-21 |
Grouping#
By “group by” we are referring to a process involving one or more of the following steps:
Splitting the data into groups based on some criteria
Applying a function to each group independently
Combining the results into a data structure
Grouping and then applying the mean() function to the resulting groups.
df.groupby("gender").mean().T
/tmp/ipykernel_1821/2544976795.py:1: FutureWarning: The default value of numeric_only in DataFrameGroupBy.mean is deprecated. In a future version, numeric_only will default to False. Either specify numeric_only or select only columns which should be valid for the function.
df.groupby("gender").mean().T
| gender | female | male |
|---|---|---|
| height | 164.363636 | 165.777778 |
| average_height_parents | 164.863636 | 165.944444 |
| number | 42.000000 | 42.000000 |
| height_m | 1.643636 | 1.657778 |
| weight | 81.221818 | 75.017778 |
| bmi | 30.050909 | 27.284444 |
Segment data into bins#
Use the function cut when you need to segment and sort data values into bins. This function is also useful for going from a continuous variable to a categorical variable.
In our example, we create a body mass index category. The standard weight status categories associated with BMI ranges for adults are shown in the following table:
BMI |
Weight Status |
|---|---|
Below 18.5 |
Underweight |
18.5 - 24.9 |
Normal or Healthy Weight |
25.0 - 29.9 |
Overweight |
30.0 and Above |
Obese |
Source: U.S. Department of Health & Human Services
In our function, we discretize the variable bmi into four bins according to the table above:
The bins [0, 18.5, 25, 30, float(‘inf’)] indicate (0,18.5], (18.5,25], (25,30], (30, float(‘inf))
float('inf')is used for setting variable with an infinitely large value
df['bmi_category'] = pd.cut(df['bmi'],
bins=[0, 18.5, 25, 30, float('inf')],
labels=['underweight', 'normal', 'overweight', "obese"])
df['bmi_category']
0 overweight
1 normal
2 overweight
3 obese
4 overweight
5 overweight
6 obese
7 overweight
8 obese
9 overweight
10 obese
11 overweight
12 obese
13 obese
14 obese
15 overweight
16 normal
17 normal
18 overweight
19 overweight
Name: bmi_category, dtype: category
Categories (4, object): ['underweight' < 'normal' < 'overweight' < 'obese']

