Generalized Additive Models (GAM)
Contents
Generalized Additive Models (GAM)#
The following code tutorial is mainly based on the statsmodels documentation about generalized additive models (GAM). To learn more about this method, review “An Introduction to Statistical Learning” from James et al. [2021]. GAMs were originally developed by Trevor Hastie and Robert Tibshirani (who are two coauthors of James et al. [2021]) to blend properties of generalized linear models with additive models.
A generalized additive model (GAM) is a way to extend the multiple linear regression model [James et al., 2021].
Remember that the basic regression model can be stated as:
which equals
In order to allow for non-linear relationships between each feature and the response we need to replace each linear component \(\beta_j x_{ij}\) with a (smooth) nonlinear function \(f_j (x_{ij})\).
We would then write the model as:
This is an example of a GAM. It is called an additive model because we calculate a separate \(f_j\) for each \(X_j\), and then add together all of their contributions.
In particular, generalized additive models allow us to use and combine regression splines, smoothing splines and local regression to deal with multiple predictors in one model. This means you can combine the different methods in your model and are able to decide which method to use for every feature.
Note that in most situations, the differences in the GAMs obtained using smoothing splines versus natural splines are small [James et al., 2021]).
In Regression splines, we discussed regression splines, which we created by specifying a set of knots, producing a sequence of basis functions, and then using least squares to estimate the spline coefficients.
In this tutorial, we use a GAM with a reguralized estimation of smooth components using B-Splines.
Note that we could also use additive smooth components using cyclic cubic regression splines (e.g. in time series data with seasonal effects).
Setup#
import numpy as np
from statsmodels.gam.api import BSplines
from statsmodels.gam.api import GLMGam
from statsmodels.tools.eval_measures import mse, rmse
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
Data preparation#
See Hitters data preparation for details about the data preprocessing steps.
We simply import the preprocessed data by using this Python script which will yield:
X_train, X_test, y_train, y_test
df_train and df_tests
from hitters_data import *
df_train
| AtBat | Hits | HmRun | Runs | RBI | Walks | Years | CAtBat | CHits | CHmRun | ... | CWalks | PutOuts | Assists | Errors | League_N | Division_W | NewLeague_N | Salary | y_pred | pred_train | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 260 | 0.644577 | 0.257439 | -0.456963 | 0.101010 | -0.763917 | -0.975959 | -0.070553 | 0.298535 | 0.239063 | -0.407836 | ... | -0.361084 | -0.482387 | 1.746229 | 3.022233 | 1 | 1 | 1 | 875.0 | 662.397284 | 662.397284 |
| 92 | -0.592807 | -0.671359 | -0.572936 | -0.778318 | -0.685806 | -0.458312 | -1.306911 | -1.001403 | -0.969702 | -0.746705 | ... | -0.844319 | -0.851547 | 0.022276 | 2.574735 | 0 | 0 | 0 | 70.0 | 106.459608 | 106.459608 |
| 137 | -0.413075 | -0.105019 | -0.688910 | -0.258715 | -0.646751 | -0.081841 | 1.577925 | 0.717456 | 0.706633 | -0.010542 | ... | 0.220252 | -0.294452 | -0.434676 | 0.635577 | 0 | 0 | 0 | 430.0 | 756.439710 | 756.439710 |
| 90 | -0.613545 | -0.558091 | 0.122907 | -0.618440 | -0.256196 | -1.211253 | -0.482672 | -0.514087 | -0.478077 | -0.501317 | ... | -0.684451 | 0.786178 | -0.545452 | -0.706917 | 0 | 1 | 0 | 431.5 | 232.771697 | 232.771697 |
| 100 | 0.637665 | 0.982354 | 0.586803 | 0.260888 | 1.227914 | 1.706394 | 0.547626 | 1.267183 | 1.437305 | 2.384908 | ... | 1.615457 | 2.504446 | -0.213124 | 0.635577 | 0 | 0 | 0 | 2460.0 | 1121.121890 | 1121.121890 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 274 | 0.824309 | 0.733164 | 0.470829 | 0.740521 | 0.954525 | 0.859335 | -0.688732 | -0.824858 | -0.808834 | -0.571428 | ... | -0.648118 | 3.427344 | 0.326910 | 1.232241 | 1 | 0 | 1 | 200.0 | 317.381231 | 317.381231 |
| 196 | 0.423369 | 0.461321 | 1.862516 | 0.500704 | 1.618469 | 0.482865 | 1.165805 | 1.354814 | 1.246368 | 1.625375 | ... | 0.681687 | -1.002566 | -0.822392 | -1.303581 | 0 | 1 | 0 | 587.5 | 936.983487 | 936.983487 |
| 159 | 1.474109 | 1.254197 | 1.746542 | 1.140215 | 2.126191 | -0.458312 | -0.894792 | -0.522636 | -0.520174 | -0.068968 | ... | -0.662651 | -0.633407 | 1.310048 | 0.933909 | 0 | 1 | 0 | 200.0 | 622.401072 | 622.401072 |
| 17 | -1.470728 | -1.396275 | -1.152806 | -1.217982 | -1.740306 | -1.258312 | -0.482672 | -0.932153 | -0.933620 | -0.770075 | ... | -0.818885 | -0.660255 | 0.403069 | 1.083075 | 0 | 1 | 0 | 175.0 | 31.929032 | 31.929032 |
| 162 | -1.643547 | -1.554850 | -1.152806 | -1.657646 | -1.701250 | -1.211253 | -0.894792 | -1.053127 | -1.020819 | -0.805130 | ... | -0.895185 | 0.111623 | -0.690846 | -1.005249 | 0 | 1 | 1 | 75.0 | 169.179139 | 169.179139 |
184 rows × 22 columns
Spline basis#
First we have to create a basis spline (“B-Spline”).
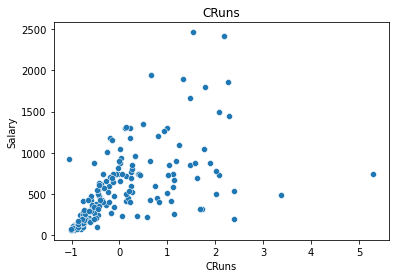
In this example, we only select two features:
CRunsandHits.
# choose features
x_spline = df_train[['CRuns', 'Hits']]
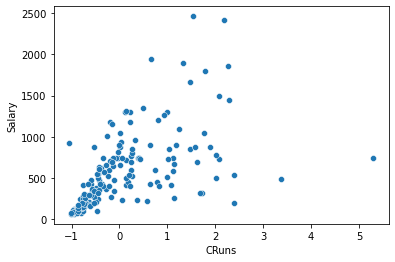
sns.scatterplot(x='CRuns', y='Salary', data=df_train);
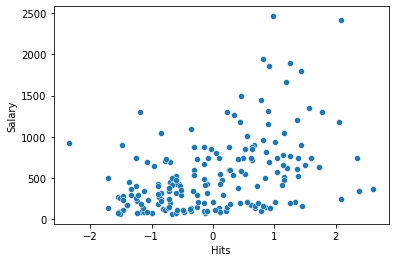
sns.scatterplot(x='Hits', y='Salary', data=df_train);
Now we need to divide the range of X into K distinct regions (for every feature).
Within each region, a polynomial function is fit to the data.
In the statsmodels function
BSplines, we need to providedfanddegree:df: number of basis functions or degrees of freedom; should be equal in length to the number of columns of x; may be an integer if x has one column or is 1-D.degree: degree(s) of the spline; the same length and type rules apply as to df.
Degrees of freedom#
Instead of providing the number of knots, in statsmodels, we have to specify the degrees of freedom (df).
dfdefines how many parameters we have to estimate.They have a specific relationship with the number of knots and the degree, which depends on the type of spline (see Stackoverflow):
In the case of B-splines:
\(df=𝑘+degree\) if you specify the knots or
\(𝑘=df−degree\) if you specify the degrees of freedom and the degree.
As an example:
A cubic spline (degree=3) with 4 knots (K=4) will have \(df=4+3=7\) degrees of freedom. If we use an intercept, we need to add an additional degree of freedom.
A cubic spline (degree=3) with 5 degrees of freedom (df=5) will have \(𝑘=5−3=2\) knots (assuming the spline has no intercept).
Note
The higher the degrees of freedom, the “wigglier” the spline gets because the number of knots is increased [James et al., 2021].
Model#
We want to fit a cubic spline (degree=3) with an intercept and three knots (K=3).
We use this values for both of our features (we also could use different values for one of the features).
This equals \(df=3+3+1=7\) for both of the features. This means that these degrees of freedom are used up by an intercept, plus six basis functions.
# create basis spline
bs = BSplines(x_spline, df=[7, 7], degree=[3, 3])
# fit model
reg = GLMGam.from_formula('Salary ~ CRuns + Hits',
data=df_train,
smoother=bs).fit()
# print results
print(reg.summary())
Generalized Linear Model Regression Results
==============================================================================
Dep. Variable: Salary No. Observations: 184
Model: GLMGam Df Residuals: 171.00
Model Family: Gaussian Df Model: 12.00
Link Function: identity Scale: 90949.
Method: PIRLS Log-Likelihood: -1304.8
Date: Sat, 23 Apr 2022 Deviance: 1.5552e+07
Time: 14:34:52 Pearson chi2: 1.56e+07
No. Iterations: 3 Pseudo R-squ. (CS): 0.7307
Covariance Type: nonrobust
==============================================================================
coef std err z P>|z| [0.025 0.975]
------------------------------------------------------------------------------
Intercept 690.7415 160.681 4.299 0.000 375.813 1005.670
CRuns 98.7724 74.429 1.327 0.184 -47.106 244.651
Hits -98.4006 112.343 -0.876 0.381 -318.588 121.787
CRuns_s0 -312.2840 407.009 -0.767 0.443 -1110.007 485.439
CRuns_s1 -206.0292 267.477 -0.770 0.441 -730.274 318.216
CRuns_s2 114.0089 284.464 0.401 0.689 -443.531 671.548
CRuns_s3 432.3174 322.183 1.342 0.180 -199.150 1063.785
CRuns_s4 208.7568 418.248 0.499 0.618 -610.994 1028.508
CRuns_s5 -216.1141 255.883 -0.845 0.398 -717.636 285.408
Hits_s0 -605.1108 497.421 -1.216 0.224 -1580.038 369.816
Hits_s1 -412.2057 280.829 -1.468 0.142 -962.620 138.208
Hits_s2 -244.4009 207.209 -1.179 0.238 -650.523 161.722
Hits_s3 109.4752 176.015 0.622 0.534 -235.508 454.458
Hits_s4 507.2235 255.672 1.984 0.047 6.116 1008.331
Hits_s5 68.8706 231.932 0.297 0.767 -385.708 523.450
==============================================================================
Plot#
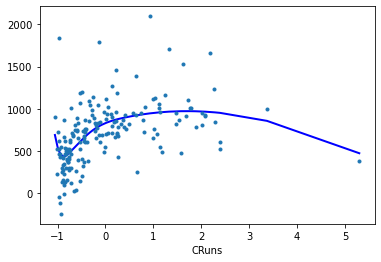
The results classes provide a
plot_partialmethod that plots the partial linear prediction of a smooth component.The partial residual or component plus residual can be added as scatter point with
cpr=True.Spline for feature
CRuns:
reg.plot_partial(0, cpr=True, plot_se=False)
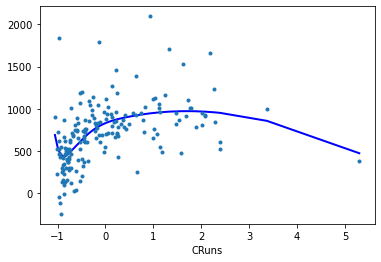
Spline for feature
Hits:
reg.plot_partial(1, cpr=True, plot_se=False)
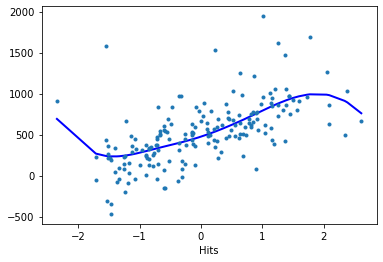
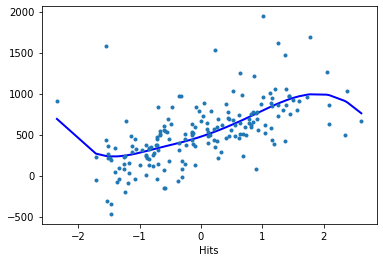
Model evaluation#
Training data#
# Training data
df_train['pred_train'] = reg.predict()
rmse_train = round(rmse(df_train['Salary'], df_train['pred_train']),4)
# plot training data
features = ['CRuns', 'Hits']
for i in features:
g = sns.scatterplot(x=i, y='Salary', data=df_train)
plt.title(i)
plt.show();
Test data#
We need to make sure that our test data falls inside the boundaries of the outermost knots (otherwise, we get an error)
# Obtain limits of training data features
x0_min = X_train['CRuns'].min()
x0_max = X_train['CRuns'].max()
x1_min = X_train['Hits'].min()
x1_max = X_train['Hits'].max()
# Only keep test data within training data limits
df_test = df_test[(df_test["CRuns"] > x0_min) & (df_test["CRuns"] < x0_max)]
df_test = df_test[(df_test["Hits"] > x1_min) & (df_test["Hits"] < x1_max)]
# plot test data
for i in features:
g = sns.scatterplot(x=i, y='Salary', data=df_test)
plt.title(i)
plt.show();
# Obtain test RMSE
df_test['pred_test'] = reg.predict(df_test, exog_smooth= df_test[['CRuns', 'Hits']])
rmse_test = round(rmse(df_test['Salary'], df_test['pred_test']),4)
# Save model results
results = {"model": "GAM", "rmse_train": rmse_train, "rmse_test": rmse_test}
results
{'model': 'GAM', 'rmse_train': 290.7294, 'rmse_test': 272.6471}